Engineering live biotherapeutic products against fungal pathogens such as Candida albicans has been suggested as a means to tackle the increasing threat of fungal infections and the development of resistance to classical antifungal treatments. One important challenge in the design of live therapeutics is to control their localization inside the human body. The specific binding capability to target organisms or tissues would greatly increase their effectiveness by increasing the local concentration of effector molecules at the site of infection. In this study, we utilized surface display of carbohydrate binding domains to enable the probiotic E. coli Nissle 1917 to adhere specifically to the pathogenic yeast Candida albicans. Binding was quantified using a newly developed method based on the automated analysis of microscopic images. In addition to a rationally selected chitin binding domain, a synthetic peptide of identical length but distinct sequence also conferred binding. Efficient binding was specific to fungal hyphae, the invasive form of C. albicans, while the yeast form, as well as abiotic cellulose and PET particles, was only weakly recognized.
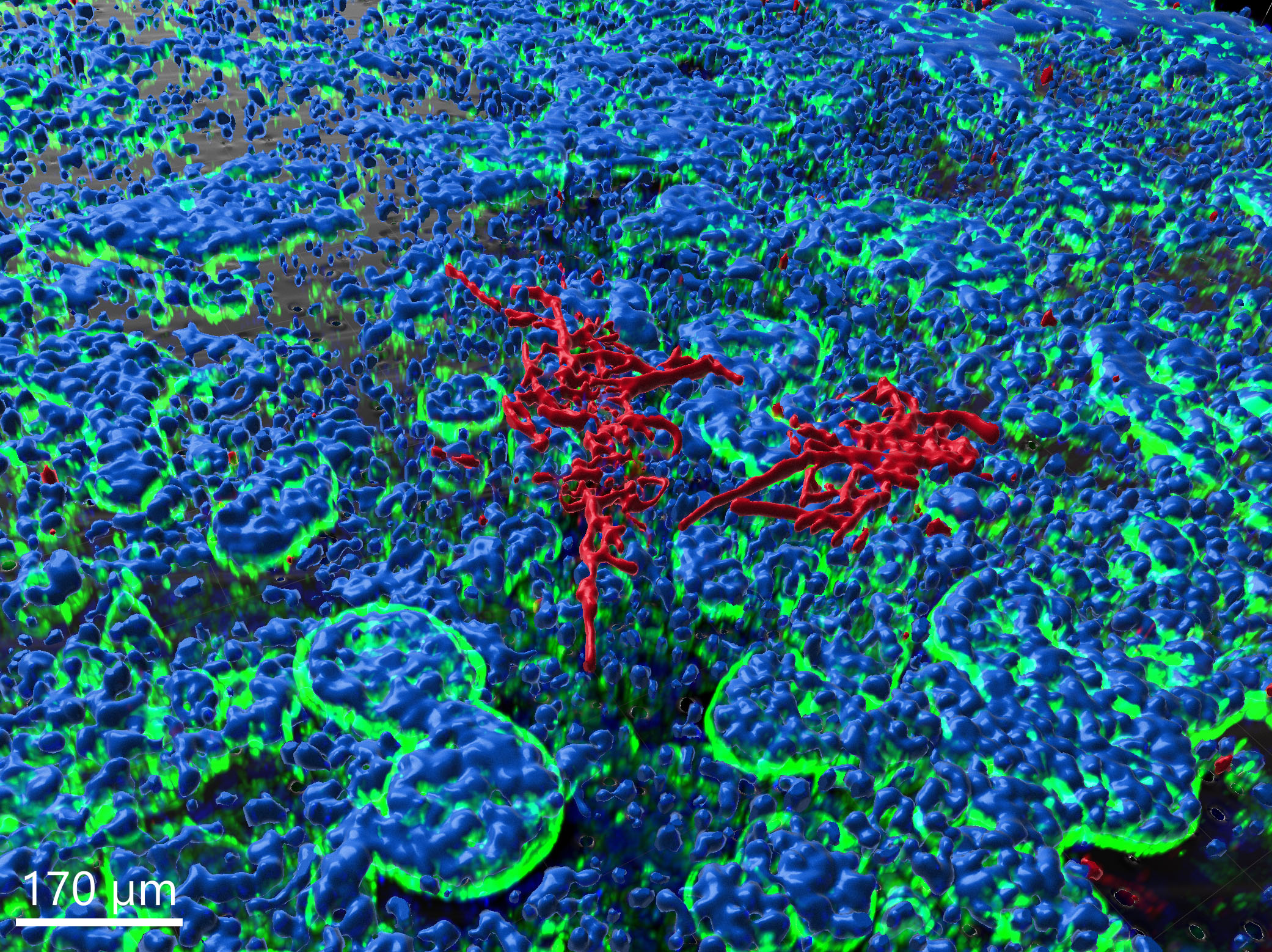
Gut-on-chip model, image-based analysis of Candida albicans
In the context of this model, the fungus is observed to proceed with hyphal growth and eventually form microcolonies.
Hyphal growth simulation
In this project, the growth of fungal hyphae is modeled. As a parameterized approach, different hyphal structures can be learned,…