Featured
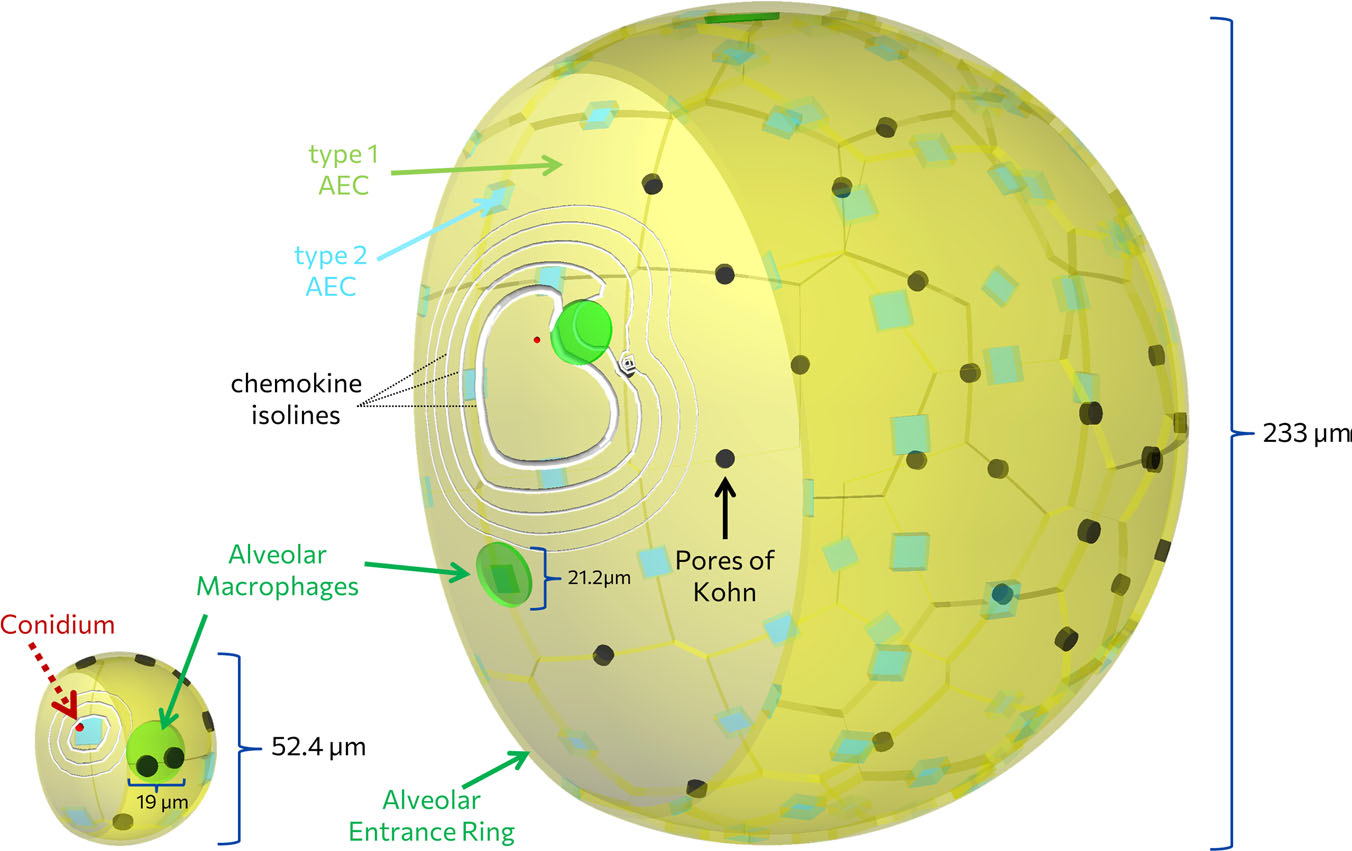
Aspergillus fumigatus infection in virtual alveolus
Lung pathogen A. fumigatus forming hyphae and macrophages eliminating the fungus, influencing infection dynamics, studied through a hybrid agent-based model.…
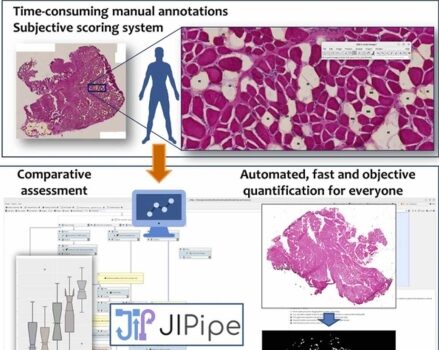
Automated Image Analysis of Liver Tissue Using JIPipe and ImageJ
Automated image analysis using JIPipe and ImageJ effectively identifies and quantifies liver tissue components in confocal images, enabling detailed studies.…
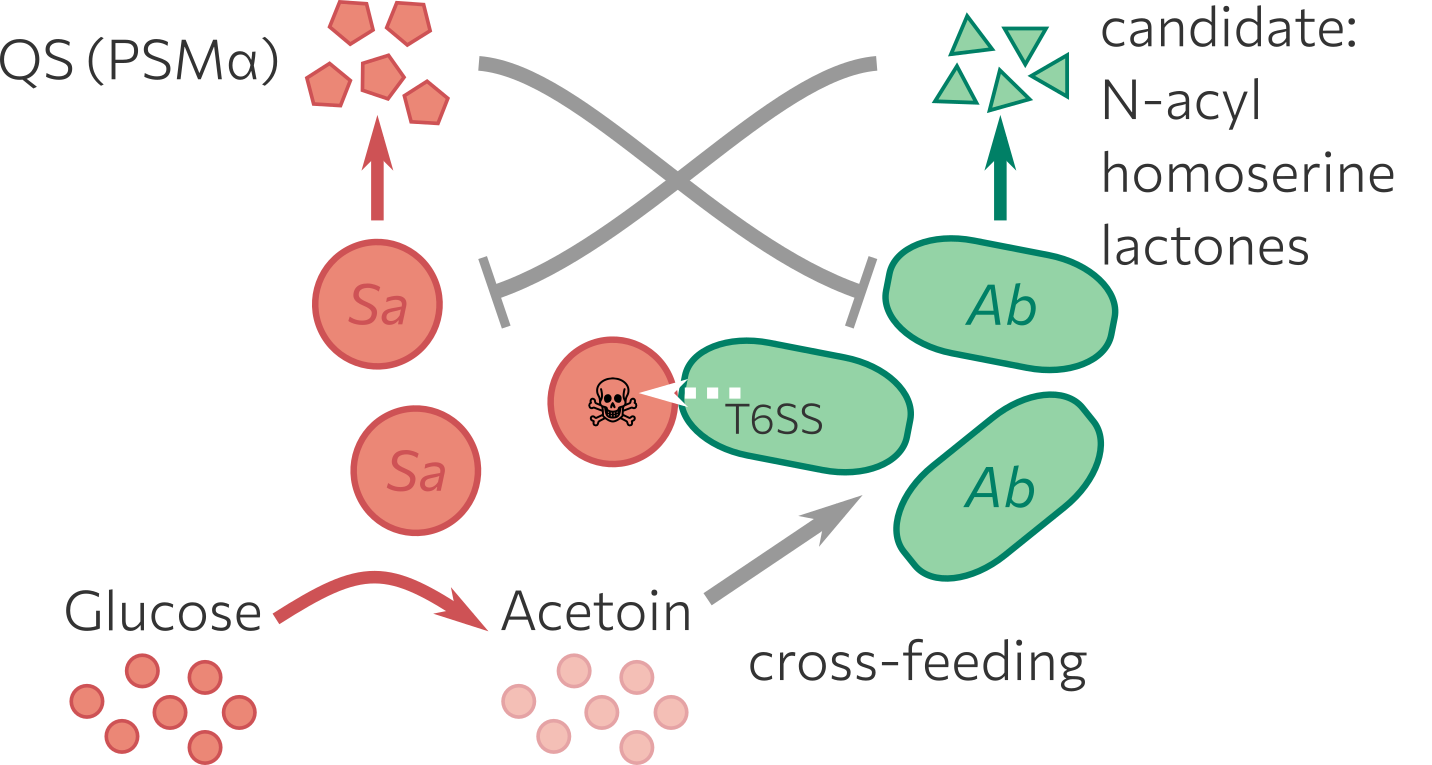
Bacterial Co-infection model
Combination of wet-lab experiments and in silico modelling to explore interactions between Staphylococcus aureus (Sa) and Acinetobacter baumannii (Ab), which…
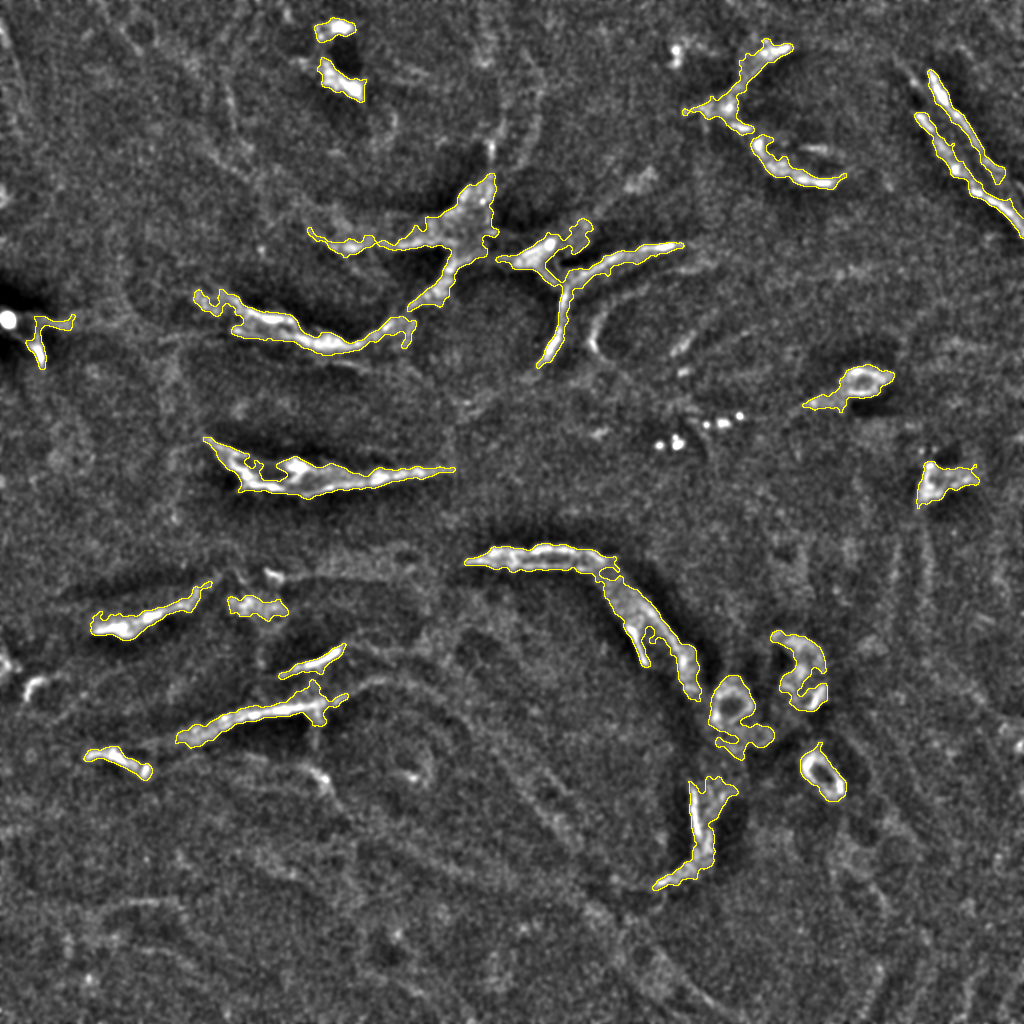
CellRain – Virtual Phagocytosis Assays
A virtual phagocytosis assay uses automated image analysis combined with individual-based modeling to simulate and uniquely characterize the phagocytosis process,…
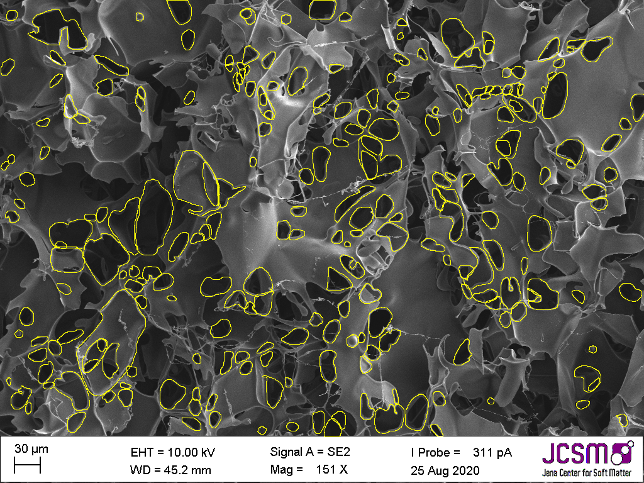
Characterizing the pore structure of cryo-polymers
A comprehensive image analysis process utilizing both classical thresholding and deep learning with Cellpose was developed to identify and measure…
Detection of Bloodstream Infections
The objective is to enhance early pathogen identification in bloodstream infections by rapid and precise analysis of neutrophil behavior to…
Droplet Analysis – Deep Learning based
This project addresses antibiotic resistance by developing a high-throughput platform for rapid antibiotic susceptibility testing.
Gut-on-chip model, image-based analysis of Candida albicans
In the context of this model, the fungus is observed to proceed with hyphal growth and eventually form microcolonies.
IMFSegNet
In this project, quantitative imaging with deep learning is employed to evaluate histological muscle samples with regard to their fat…
JIPipe
JIPipe introduces a visual programming language into ImageJ that allows the creation of pipelines by designing flow charts. This language…
KITE – EPIDEMIOLOGICAL MODELLING OF INFECTION SPREAD IN CHILD CARE FACILITIES
KITE (KIndergarten Test scenario Evaluator) is a state-based model for simulating the spread of different airborne viruses in kindergartens. The…
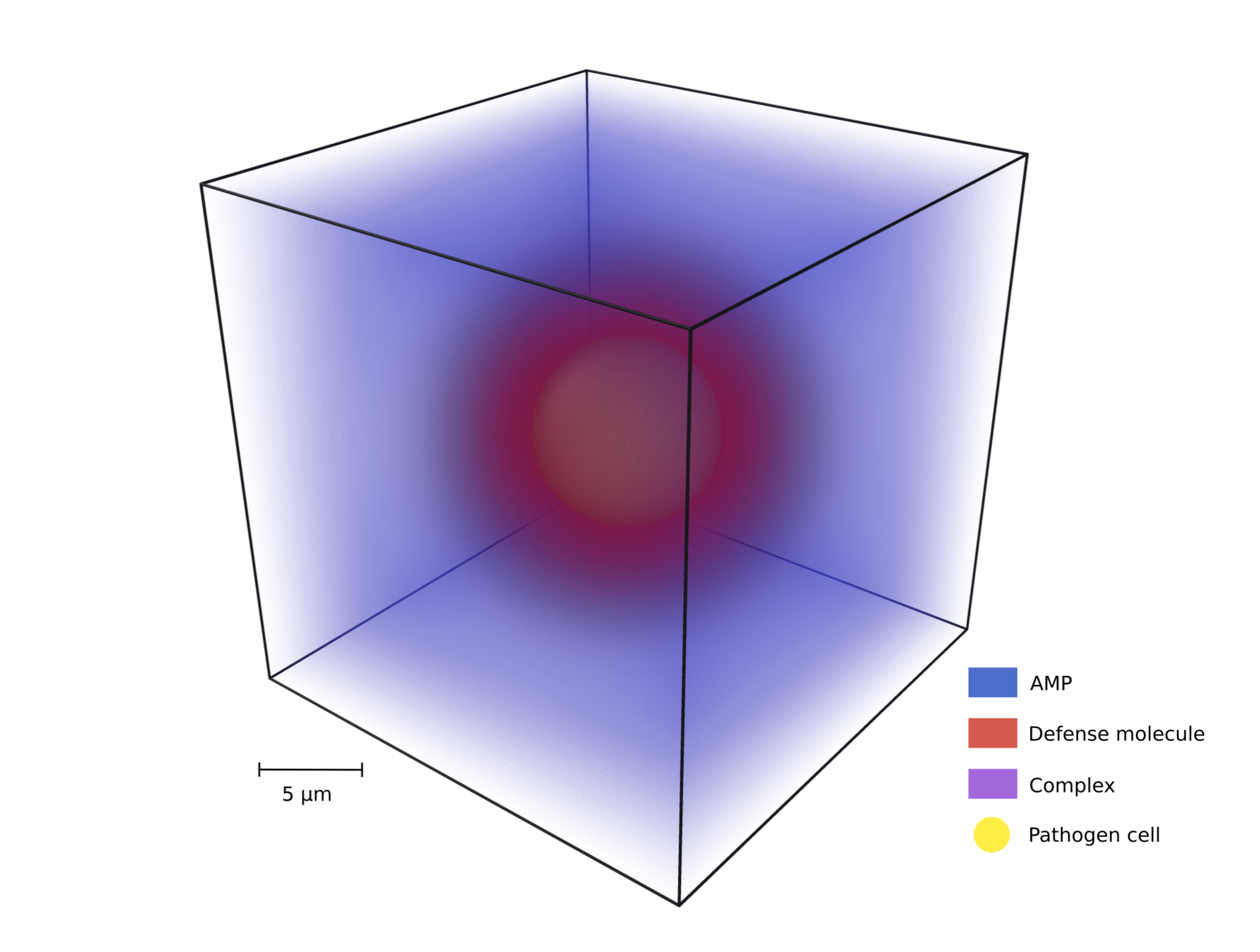
Modeling pathogen AMP evasion
This project models the antimicrobial peptide (AMP) evasion mechanism called spatial distancing, where pathogens secrete molecules that bind AMPs and…
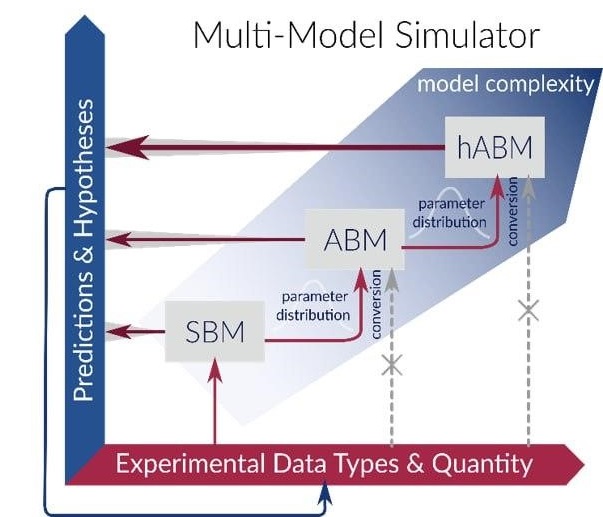
Mu Mo Sim
The Multi-Model Simulator project introduces a novel simulation tool that integrates various modeling approaches to provide reliable quantitative predictions with…
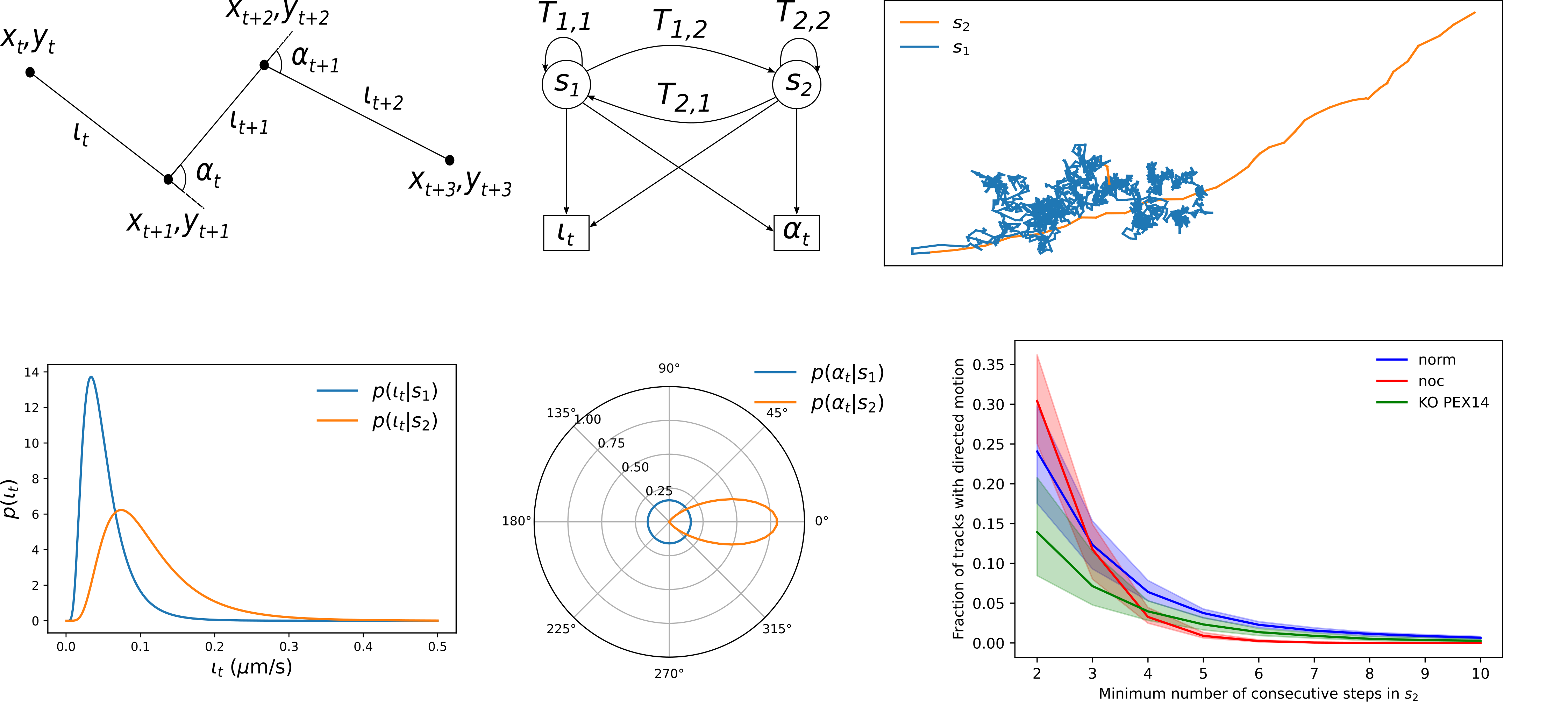
Peroxisome motility analysis
Peroxisome motility analysis using a Hidden Markov Model (HMM) enables the automatic identification and quantification of migration segments, thereby enhancing…
All projects
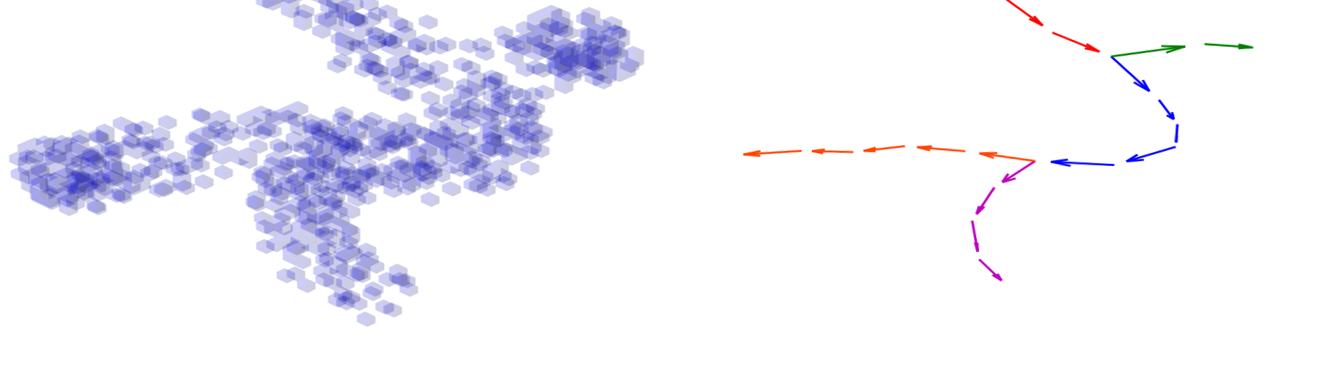
3D-HyTracer
3D-HyTracer is an image analysis tool for automated tracing of fungal hyphae. It creates a low-memory and easily accessible spatial…
ABM
The agent-based modelling framework is a generalized framework that is able to capture spatio-temporal dynamics of processes in host-pathogen interactions…
ACAQ
ACAQ is an ImageJ macro designed for the analysis of phagocytosis assays. ACAQ was gradually developed into a full-blown image…
AMIT
The algorithm for migration and interaction tracking (AMIT) approach enables high-throughput segmentation for label-free cells, which does not rely on…
Aspergillus fumigatus – macrophage interaction
The first line of defence against the airborne human-pathogenic fungus A. fumigatus are the resident immune cells in the human…
Aspergillus fumigatus infection in virtual alveolus
Lung pathogen A. fumigatus forming hyphae and macrophages eliminating the fungus, influencing infection dynamics, studied through a hybrid agent-based model.…
Automated Image Analysis of Liver Tissue Using JIPipe and ImageJ
Automated image analysis using JIPipe and ImageJ effectively identifies and quantifies liver tissue components in confocal images, enabling detailed studies.…
Bacterial Co-infection model
Combination of wet-lab experiments and in silico modelling to explore interactions between Staphylococcus aureus (Sa) and Acinetobacter baumannii (Ab), which…