Although multispectral optoacoustic tomography (MSOT) significantly evolved over the last several years, there is a lack of quantitative methods for analysing this type of image data. Current analytical methods characterise the MSOT signal in manually defined regions of interest outlining selected tissue areas. These methods demand expert knowledge of the sample anatomy, are time consuming, highly subjective and prone to user bias. Here we present our fully automated open-source MSOT cluster analysis toolkit Mcat that was designed to overcome these shortcomings. It employs a deep learning-based approach for initial image segmentation followed by unsupervised machine learning to identify regions of similar signal kinetics. It provides an objective and automated approach to quantify the pharmacokinetics and extract the biodistribution of biomarkers from MSOT data. We exemplify our generally applicable analysis method by quantifying liver function in a preclinical sepsis model whilst highlighting the advantages of our new approach compared to the severe limitations of existing analysis procedures.
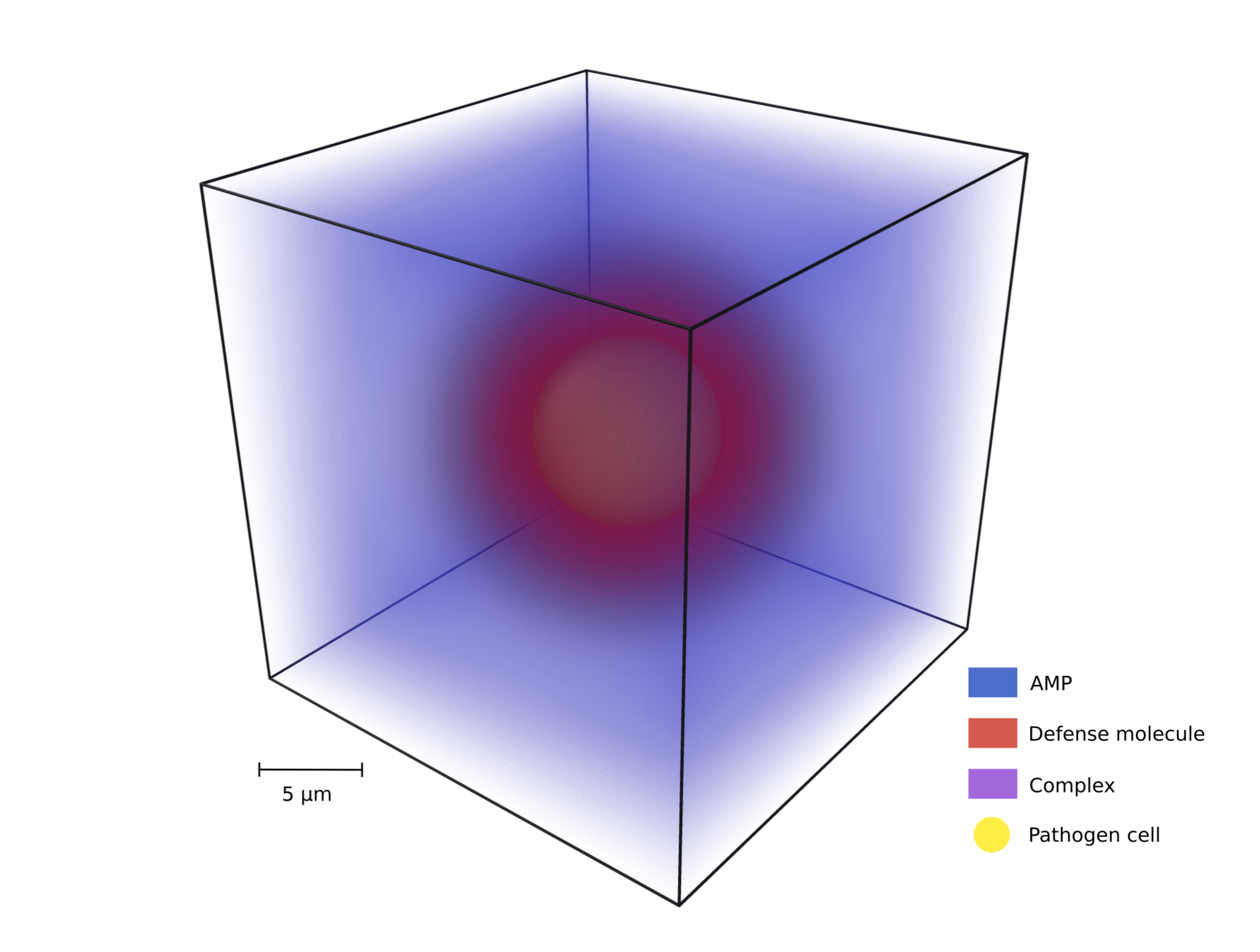
Modeling pathogen AMP evasion
This project models the antimicrobial peptide (AMP) evasion mechanism called spatial distancing, where pathogens secrete molecules that bind AMPs and…
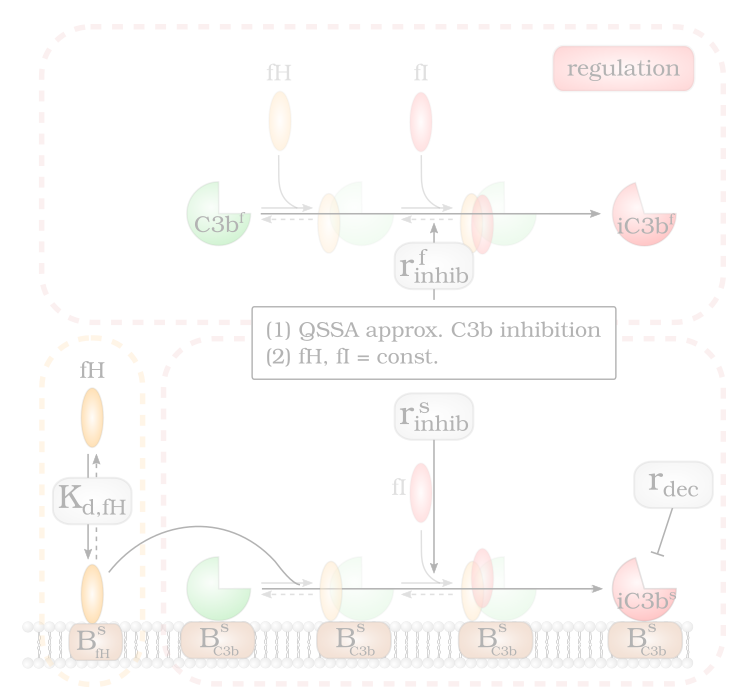
Modelling Factor H Mediated Self vs. Non-self Discrimination
By screening the Factor H concentration on cell surfaces three opsonization regimes are identified: (1) non-self recognition, (2) complement evasion…