Hyphal growth segmentation
Hyphal growth is the driving force of fungal bloodstream infections due to their ability to pierce through epithelial tissue. Investigating the growth of a hypha via image analysis is often hindered by crossing or touching hyphae from different fungal branches and the quality of the image. The 3D-HyTracer can, to some extent, overcome common problems resulting from, e.g., regional lack of signal, low signal-to-noise ratio, or crowded hyphal structures. It can handle gaps in disrupted hyphal segmentations and detect and correct atypical hyphal traces for the underlying segmentation. Similarly, hyphal structures originating from different spores can be dissected by the use of 3D-HyTracer.
Experimental Collaborators
- Infections in Hematology/Oncology group at the Leibniz-HKI in Jena, Germany.
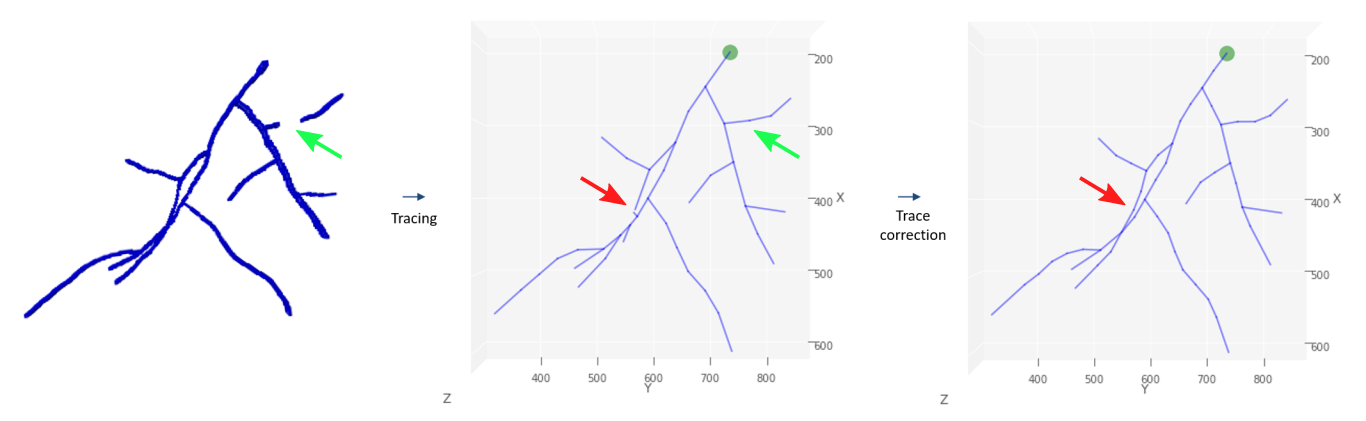
Visualized processing steps of the 3D-HyTracer on simulated hyphae
From left to right, we have a segmented hyphal structure that is then traced using 3D-HyTracer in the middle panel. Not how a part of hyphae is missing at the green arrow in the left-most image, and this is corrected in the tracing step; see green arrow in the middle panel. In the middle and right-most panel, red arrows, we see how errors are corrected when a mistake in tracing occurred by taking the most probable branching angles and structures.
Technical details
The 3D-Hytracer creates a low-memory and easily accessible spatial graph in NetworkX format for given hyphal segmentations. The very core functionality of the 3D-HyTracer is based on steepest descent foreground tracing, a method adapted from the neuron tracing tool Rivulet2. It requires a scanned hyphal structure and the positions of fungal spores. In preparation for the tracing, the 3D-Hytracer assigns every voxel with a cost value based on the distance transform of the binarized initial image and a distance value, equating values on the shortest path between the voxel and the spore in the cost map. The underlying shortest-path problem is solved by approximating the Eikonal equation using the marching cubes algorithm. The resulting distance values allow the hyphal tracing even on poorly segmented images. This is done for every spore individually before the automated correction of hyphal traces. The correction of hyphal traces is done using known properties of hyphal growth, e.g. the calculation of branching rates and angles, turning angles, hyphal orientation, length, and diameter. The correction step of hyphal tracing can readily be adjusted for the entire graph or parts of the graph if, for example, a different fungus with different branching patterns is analyzed. In fungal pellets, a collection of clustered spores with a dense core of inseparable hyphae, fully tracing a hypha back to the corresponding spore might not be possible. To enable an analysis of such samples, the 3D-Hytracer is extended by a module that first distinguishes between pellet core and periphery and then traces/analyses the hyphal periphery only.




