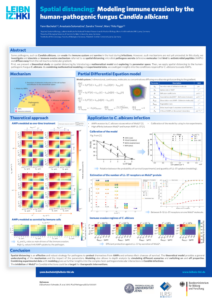
The agent-based modelling framework is able to capture spatio-temporal dynamics of processes in pathogen-host interactions in the human system. This approach provides a variety of possibilities, as it can represent the specific morphology of cells as well as their migration and interactions within a defined environment. Although ABMs are very flexible and can be used for almost every biological system, their stochastic nature makes them computationally expensive. This is especially true if one wants to combine them with partial differential equations (PDE) to also mimic molecule-cell interactions. Therefore, we developed an efficient and scaleable framework in C++ to simulate complex models. This framework has been applied vor various studies to investigate host-pathogen interactions in whole-blood, the lung or to investigate antibody-antigen binding (for publications see below).
Features
- Completly developed in modern C++
- Parallelized with openMP
- Integration of high-effecient PDE solvers to simulate molecule and cell interactions
Publications
- Lehnert, Leonhardt et al. (2021) Ex vivo immune profiling in patient blood enables quantification of innate immune effector functions. Scientific Reports 11, 12039.
- Blickensdorf et al. (2020). Hybrid Agent-Based Modeling of Aspergillus fumigatus Infection to Quantitatively Investigate the Role of Pores of Kohn in Human Alveoli. Frontiers in Microbiology, 11(Aug), 1–13.
- Blickensdorf et al. (2019). Comparative assessment of aspergillosis by virtual infection modeling in murine and human lung. Frontiers in Immunology, 10(Feb).
- Timme et al. (2018) Quantitative simulations predict treatment strategies against fungal infections in virtual neutropenic patients. Frontiers in Immunology 9, 667.
- Lehnert et al. (2017) Dimensionality of motion and binding valency govern receptor-ligand kinetics as revealed by agent-based modeling. Frontiers in Immunology 8, 1692.
- Lehnert, Timme et al. (2015) Bottom-up modeling approach for the quantitative estimation of parameters in pathogen-host interactions. Frontiers in Microbiology 6(608).
- Pollmächer et al. (2015). Deciphering chemokine properties by a hybrid agent-based model of Aspergillus fumigatus infection in human alveoli. Frontiers in Microbiology, 6(May), 1–14.
- Pollmächer et al. (2014). Agent-based model of human alveoli predicts chemotactic signaling by epithelial cells during early Aspergillus fumigatus infection. PLoS ONE, 9(10).