Staphylococcus aureus (Sa) and Acinetobacter baumannii (Ab) are often found together in severe polymicrobial infections resistant to treatment. In this project a combination of wet-lab experiments and in silico modeling was used to explore the interactions, using both, representative laboratory strains and strains co-isolated from clinical samples.
The in silico model developed quantifies the interaction dynamics between Ab and Sa using ordinary differential equations (ODEs).
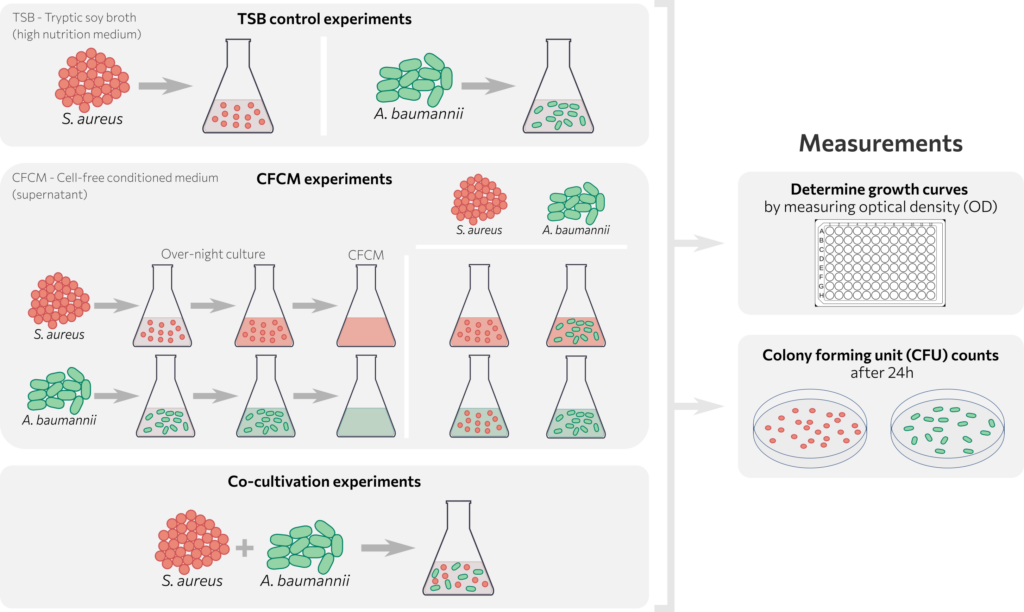
This model simulates bacterial growth in monocultures and co-cultures, modelling population dynamics with a logistic growth model. The model includes a time-dependent carrying capacity to reflect changes in growth conditions over time, such as nutrient availability. Numerical simulations were conducted using the Python programming language with the SciPy.optimize package, integrating ODEs based on experimental initial conditions. Model parameters were estimated by fitting simulations to experimental data using the simulated annealing method, with the objective of optimizing parameters to minimize the sum of the squares of the error between simulated and experimental growth curves.
Both typical lab strains and strains isolated from clinical samples were analyzed. We found that Sa can partially interfere with Ab through the expression of phenol-soluble modulins. Additionally, cross-feeding mechanism where Sa supports Ab growth by providing acetoin as a carbon source were identified. This research highlights the complex interplay of competitive and cooperative strategies between Ab and Sa. By shedding light on these dynamics, the study enhances our understanding of how these pathogens coexist in challenging infections, potentially paving the way for new therapeutic strategies.
Experimental Collaborators
- Tuchscherr Lab at the University Hospital Jena, Germany





