The prevention of sepsis requires the early detection of pathogens in bloodstream infections. It is known that neutrophils exhibit characteristic behaviors when infected with Candida species. In this study, blood samples from healthy donors were infected with Candida albicans and Candida glabrata, and neutrophils were imaged. This project applied advanced deep learning methods, including gated recurrent unit (GRU) networks, transformer-based networks, and convolutional neural networks (CNNs), to classify infections. Two distinct workflows were employed, one for whole image morphodynamics and another for image classification.
The objective is to enhance early pathogen identification in bloodstream infections by rapid and precise analysis of neutrophil behavior to prevent sepsis.
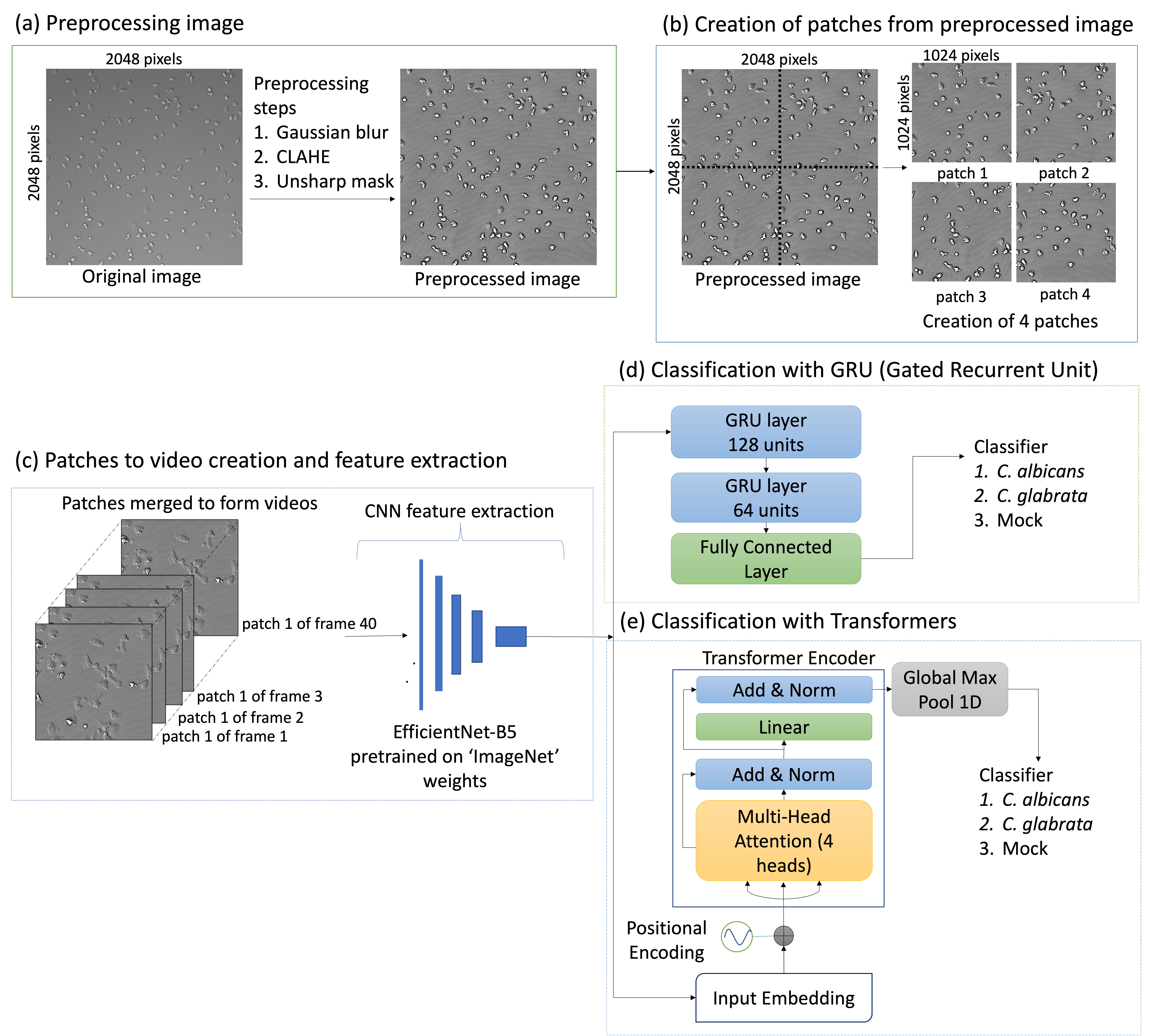
For whole image morphodynamics, video frames were preprocessed, split into patches, and compiled into videos. EfficientNetB5 extracted features, and GRUs or transformers classified the infection scenarios.
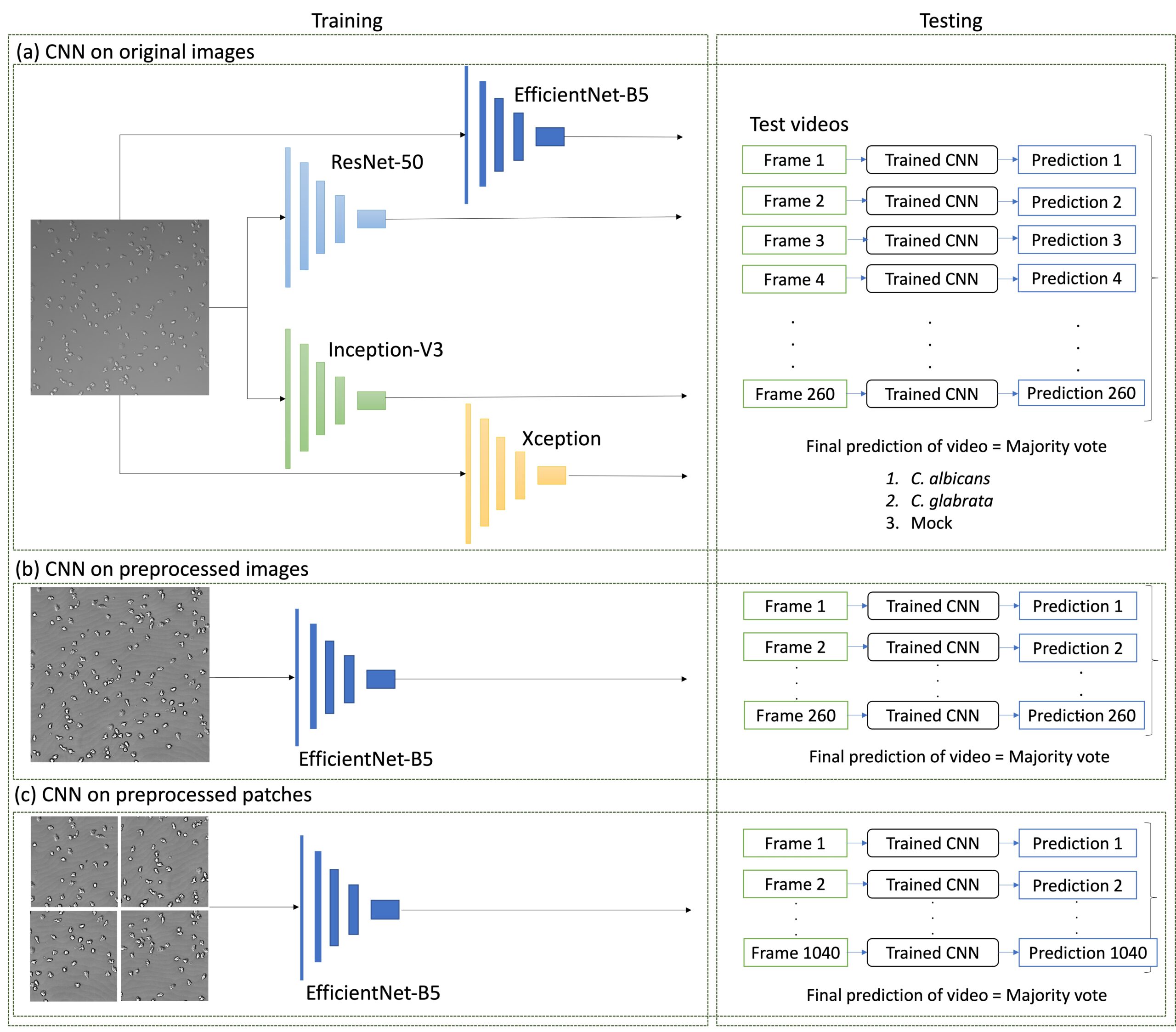
For image classification, ResNet50, Inception-V3, Xception, and EfficientNetB5 were trained on images without preprocessing, with EfficientNetB5 performing the best. During the testing phase, individual frames were classified using a majority voting technique, and EfficientNetB5 was employed for subsequent tests on preprocessed whole images and patches.
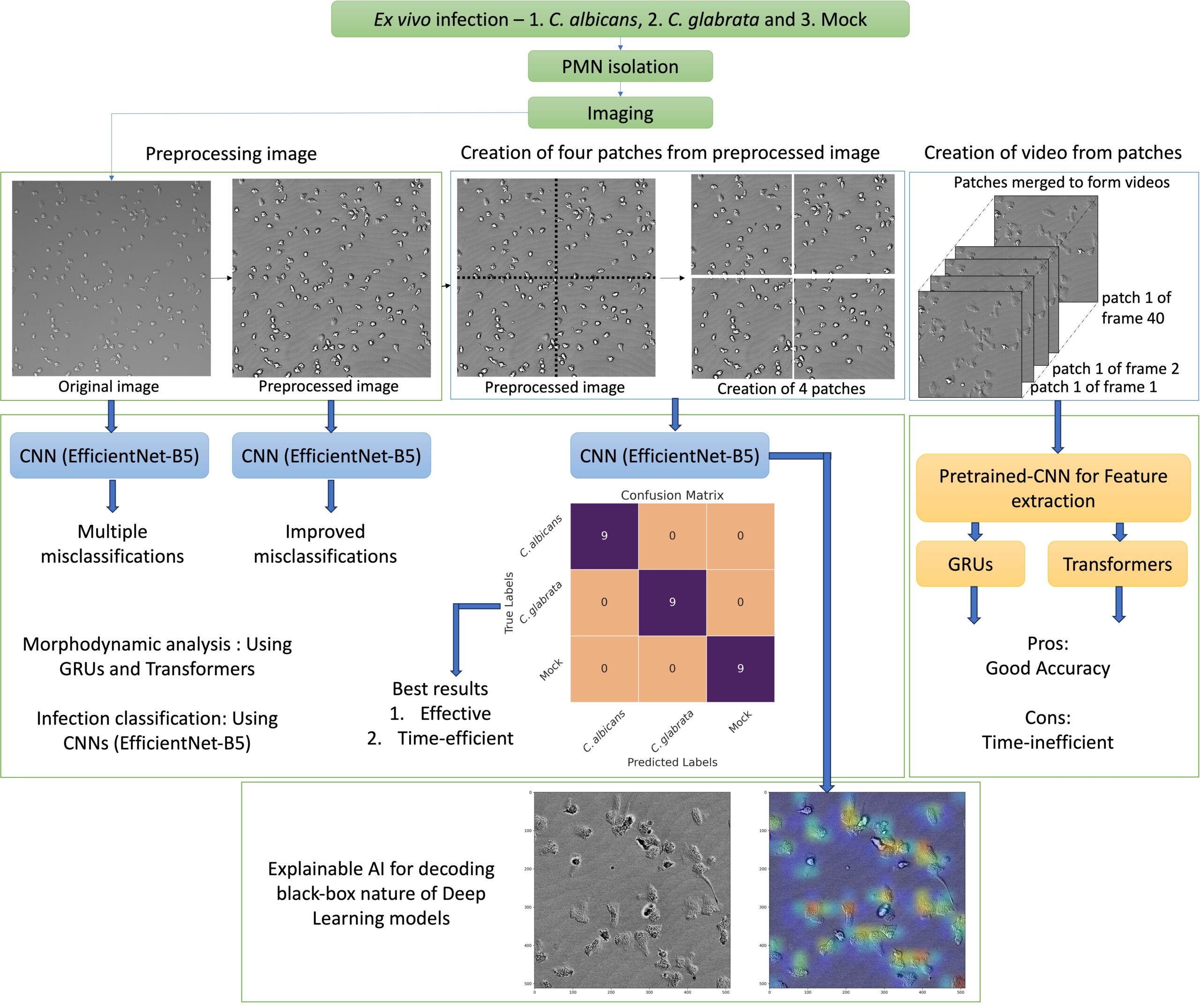
The CNN model achieved 100% accuracy with fast inference times. This approach could facilitate the development of rapid automated diagnostic tools for fungal bloodstream infections, offering a promising method for clinical settings. GRU networks achieved 96% accuracy, transformers reached 100%, and CNNs also reached 100% with faster inference times. Explainable AI techniques highlighted the importance of neutrophil morphodynamics in differentiation. CNN models can rapidly and accurately diagnose fungal bloodstream infections, offering a promising method for automated diagnostics in clinical settings. The project demonstrated that using a CNN model is effective and efficient in diagnosing fungal bloodstream infections based on neutrophil behavior. This represents a novel approach to the rapid and efficient detection of sepsis.
Experimental Collaborators
- Fungal Septomics group at the Leibniz-HKI in Jena, Germany
- Institute for Hygiene and Microbiology at the Julius-Maximilians University Würzburg in Jena, Germany





