The DynaCoSys model is a hybrid differential equation model that describes the complement activation of the alternative pathway. The dynamics of the surface bound molecules are described by a set of ordinary equations and the dynamics of the key fluid phase molecules are described by partial differential equations. The non-linear model includes all features of the complement system: activation, opsonization, stabilization, amplification and regulation. The DynaCoSys model is used in our studies for analyzing the complement activation mediated by Factor H and researching the complement immune evasion of candida albicans in the complement immune evasion study.
Hybrid differential equation model of the complement system
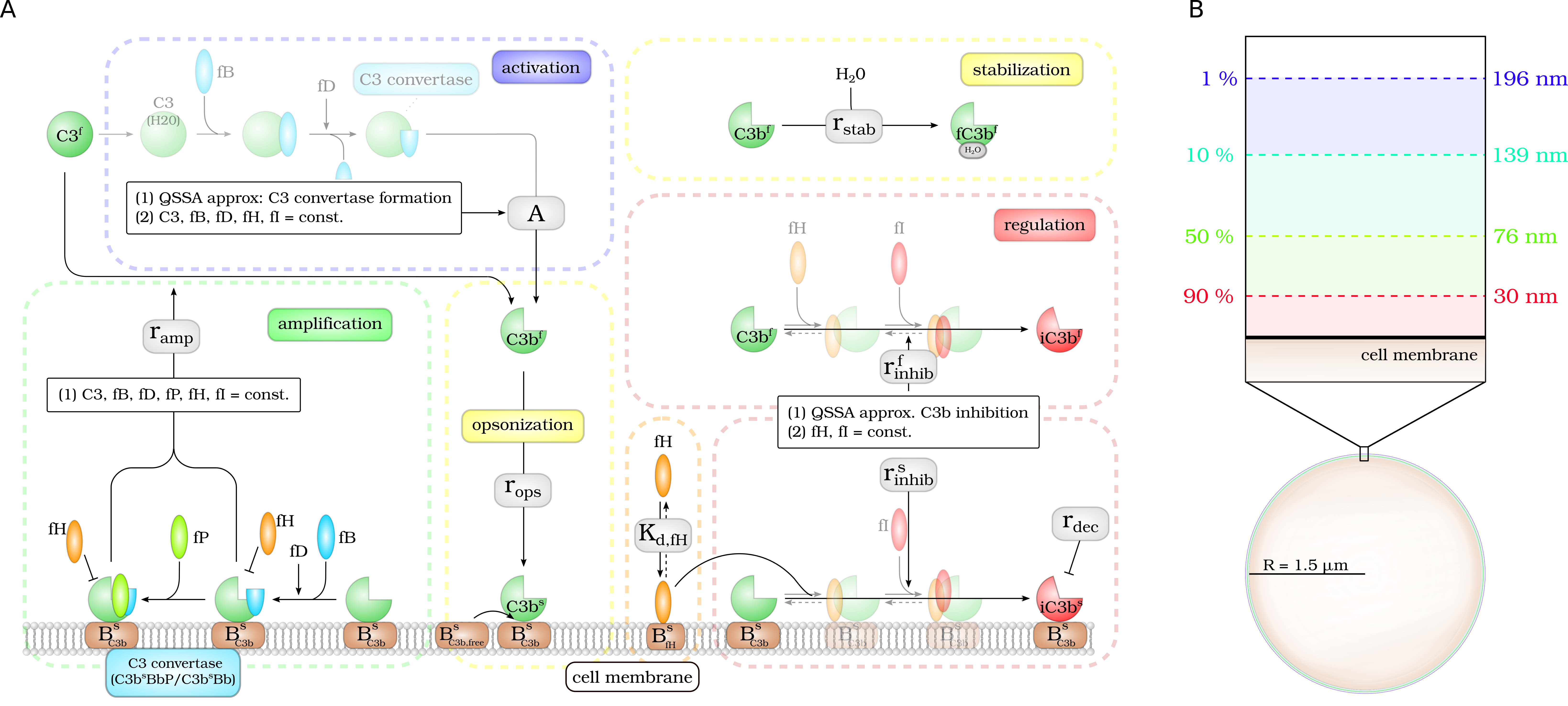
Complement activation can be divided into five parts: (i) activation, (ii) opsonization, (iii) stabilization, (iv) amplification, and (v) regulation. (A) The model focuses on the dynamics of the central component $C_3b$: Active $C_3b$ in the fluid phase, $C_3b^f$, results from cleavage of precursor molecule $C_3^f$. The interaction of the fluid phase molecule $C_3b^f$ with the cell surface is modeled by the interaction with free surface binding sites $B^s_{C_3b,free}$ and binding sites BsC3b that are occupied with molecules $C^3b_s$ on the surface. $C_3b^f$ that does not bind to the cell surface gets inactivated via a Factor H mediated inhibition process, or gets stabilized by water molecules and is no longer able to bind to the cell surface. Surface-bound $C_3b^s$ can form $C_3$-convertase molecules—$C_3b^sBb$ and $C_3b^sBbP$–that cleave $C_3^f$ molecules to $C_3b^f$ molecules in the vicinity of the cell surface. $C_3b^s$ can be inactivated via an inhibition process that is mediated by surface-bound Factor H, whose concentration depends on the concentration of binding sites on the cell surface $B^s_{fH,max}$. (B) The lifetime of active $C_3b^f$ is short such that, depending on the distance from the cell surface, the fraction of molecules that reach the cell surface is small; for example, only 1% at a distance of 196 nm within a simple decay model.






