Light-sheet fluorescence microscopy (LSFM) allows to capture whole organs in three spatial dimensions. Manual analysis of these big-volume data sets of up to hundreds of gigabytes is not feasible. Therefore, fully automated solutions need to be developed.
LSFM data of a whole murine kidney
The glomeruli are visible as bright red dots within the tissue. Manual quantification and extraction of morphological properties is not feasible.
Experimental Collaborators
- Gunzer Lab, Institute for Experimental Immunology and Imaging, University hospital Essen, Germany
- Coldewey Lab, University hospital Jena, Germany
Fully-automated analysis
The fully automated analysis was developed in Python by utilizing the popular scikit-image, Mahotas, and Numpy libraries.
Quantification of the LSFM data
The fully automated quantification tool was able to count all glomeruli in the whole kidney in 3D.
Performance improvements
Later, we converted the Python code into a high-performance C++ application via our MISA++ library. We could observe a performance increase by approximately two orders of magnitude – reducing the analysis time from about three days to only 40 minutes.
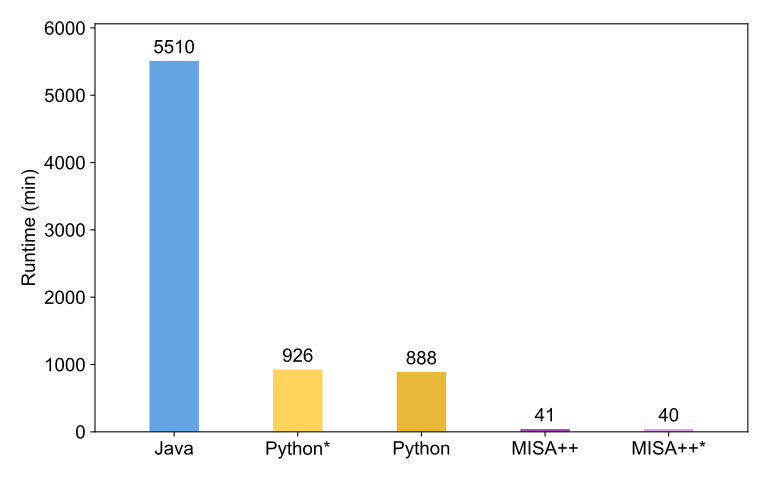
Performance comparison
We compared the performance of the glomeruli analysis workflow between the original implementation (Python), MISA++, and an ImageJ2-based Java implementation. MISA++ is the fasted tool by a large margin. Asterisks denote disabled performance optimizations.





