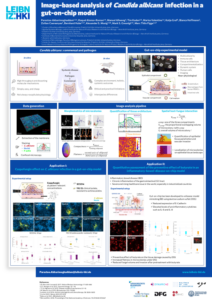
Microbial dysbiosis is closely associated with chronic inflammation and inflammatory bowel disease. Traditional in vitro models proposed to study the underlying mechanisms lack the required tissue complexity and therefore often fail to accurately reproduce the complex, multi-component structure of the intestinal wall. On the other hand, animal models are not always transferable to the human condition, as there are significant differences in the immune system and microbiota between humans and animals such as mice. This is where the three-dimensional gut-on-chip model becomes a valuable tool. The gut-on-chip model comprises three main components: a bloodstream-like compartment with endothelial cells, an intestinal epithelial barrier with resident innate immune cells and a perfused three-dimensional tissue structure of villi and crypts. An image-based approach is employed to analyse colonisation and infection with the opportunistic pathogenic fungus Candida albicans in the gut-on-chip model.
In the context of this model, the fungus is observed to proceed with hyphal growth and eventually form microcolonies.

Confocal laser scanning microscopy is employed to obtain three-dimensional multi-channel images. An automated image analysis pipeline has been developed to enable three-dimensional geometric and morphological characterisation of microcolonies. Furthermore, the complexity of microcolony surfaces is quantified using the concept of fractal dimension. A three-dimensional multilocalisation analysis is performed to quantify the interaction as well as the extent of fungal invasion into the endothelial compartment. The image depicts C. albicans in red and the nuclei of epithelial cells in blue. The combination of the gut chip model with this image-based approach enables the quantitative investigation of C. albicans infection patterns in a complex, in vivo-like environment.
Experimental Collaborators
- Center for Sepsis Control and Care, in Jena, Germany
- Microbial Pathogenicity Mechanisms at the Leibniz-HKI in Jena, Germany