In this study we investigate receptor–ligand binding in the context of antibody–antigen binding. We established a quantitative mapping between macroscopic binding rates of a deterministic differential equation model and their microscopic equivalents as obtained from simulating the spatiotemporal binding kinetics by a stochastic agent-based model. Furthermore, various properties of B cell-derived receptors like their dimensionality of motion, morphology, and binding valency are considered and their impact on receptor–ligand binding kinetics is investigated. The different morphologies of B cell-derived receptors include simple sperical representations as well as more realistic Y-shaped morphologies. These receptors move in different dimensionalities, i.e. either as membrane-anchored receptors or as soluble antibodies. The mapping of the macroscopic and microscopic binding rates allowed us to quantitatively compare different agent-based model variants for the different types of B cell-derived receptors. Our results indicate that the dimensionality of motion governs the binding kinetics and that this predominant impact is quantitatively compensated by the bivalency of these receptors.
Model for antigen binding by B cell-derived receptors
Publications
Selective activation of dynamics in kinetically frozen supramolecular polymer bottlebrush assemblies
Hans F. Ulricha, b, Tobias Kleina,b, Ziliang Zhaoc, d, Zoltán Cseresnyése, Pablo Carravillac, d, Ruman Gerste, Alina Kasbergg, Frederic P. Scharfenberga, Marc Thilo Figgee f, Christian Eggelingb, c, d, Johannes C. Brendela, b, g, *
a Laboratory of Organic and Macromolecular Chemistry (IOMC), Friedrich-Schiller-University Jena, Humboldtstraße 10, 07743 Jena, Germany.
b Jena Center for Soft Matter (JCSM), Friedrich-Schiller-University Jena, Philosophenweg 7, 07743 Jena, Germany.
c Institute for Applied Optics and Biophysics, Friedrich Schiller University Jena, 07743 Jena, Germany.
d Leibniz Institute of Photonic Technology e.V., member of the Leibniz Centre for Photonics in Infection Research (LPI), 07743 Jena, Germany.
e Applied Systems Biology, Leibniz Institute for Natural Product Research and Infection Biology, Hans Knöll Institute (HKI), 07743 Jena, Germany.
f Institute of Microbiology, Faculty of Biological Sciences, Friedrich-Schiller-University Jena, 07743 Jena, Germany.
g Macromolecular Chemistry I, University of Bayreuth, Universitätsstr. 30, 95447 Bayreuth, Germany.
Supramolecular assemblies are typically characterized by their dynamic nature due to the comparable weak non-covalentinteractions. While these properties confer adaptability, stability issues may limit application in areas such as drug delivery ortissue engineering. Here, we show that supramolecular assemblies of amphiphilic polymers containing benzenetrispeptide andbenzenetrisureas motifs are inherently stable and non-dynamic at ambient conditions, but […]
Complex-mediated evasion: modeling defense against antimicrobial peptides with application to human-pathogenic fungus Candida albicans
Yann Bachelot, Anastasia Solomatina & Marc Thilo Figge#
Understanding the complex interplay between host and pathogen during infection is critical for developing diagnostics and improving therapeutic interventions. Among the diverse arsenal employed by the host, antimicrobial peptides (AMP) play a key role in the defense against pathogens. We propose an immune evasion mechanism termed “Complex-mediated evasion” (CME), that allows pathogens to protect themselves […]
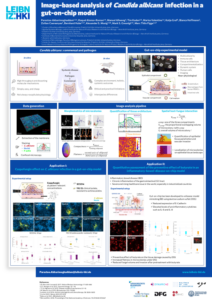
Image-based quantification of Candida albicans filamentation and hyphal length using the open-source visual programming language JIPipe
Jan-Philipp Praetorius*, Sophia U. J. Hitzler*, Mark S. Gresnigt#, Marc Thilo Figge#
The formation of hyphae is one of the most crucial virulence traits the human pathogenic fungus Candida albicans possesses. The assessment of hyphal length in response to various stimuli, such as exposure to human serum, provides valuable insights into the adaptation strategies of C. albicans to the host environment. Despite the increasing high-throughput capacity live-cell imaging and data generation, […]