In this project, an antimicrobial peptide (AMP) evasion mechanism is modeled and simulated. This mechanism, which we refer to as spatial distancing, consists of a pathogen cell secreting defense molecules that can bind to AMP and form complexes with the highest concentration close to the cell surface that diffuse away from the pathogen. As a result, AMP are transported away from the pathogen’s surface, effectively protecting it from the immune attack by AMP.
The modeling of spatial distancing, where pathogens secrete defense molecules that bind AMPs, resulting in the pathogen evading the immune system.
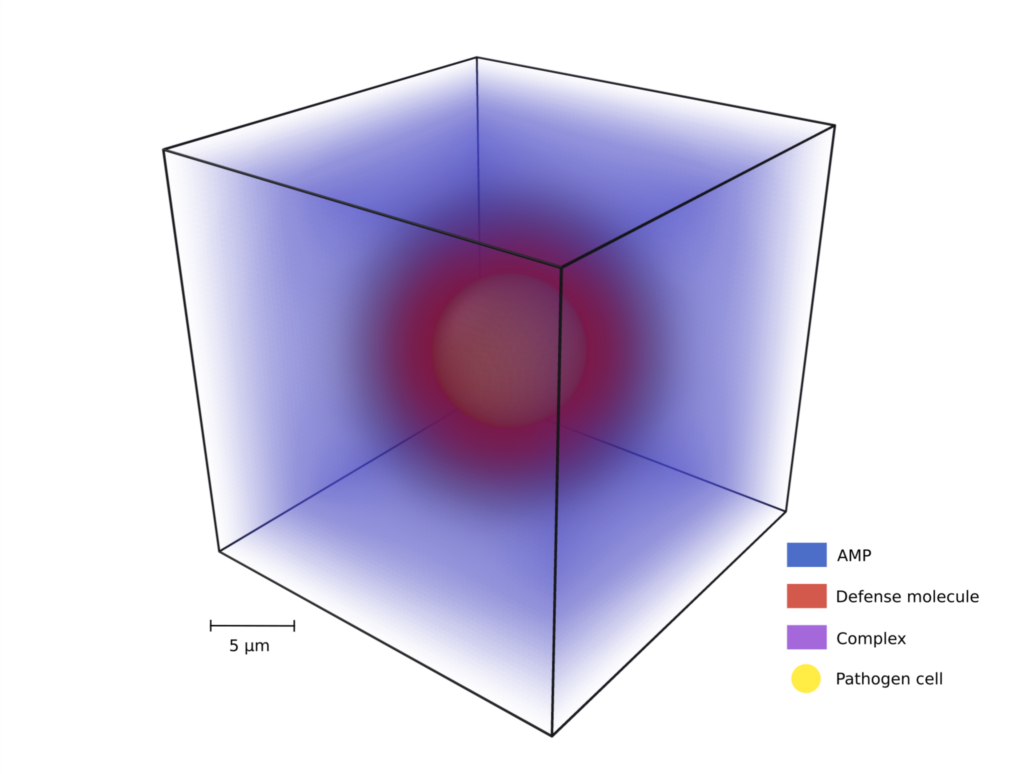
To gain a comprehensive understanding of spatial distancing, we simulate a pathogenic cell in a three-dimensional environment with molecule concentrations modeled by partial differential equations. Two different scenarios are simulated, respectively, representing AMPs secreted by immune cells and AMPs administered for medical treatment. Spatial distancing was then applied and studied in the case of the human-pathogenic fungus Candida albicans.





