Diffusion and mobility are essential for cellular functions, as they facilitate the meeting of molecules and organelles, thereby enabling them to interact and perform their respective roles within the cell. To observe these dynamics, it is necessary to have high spatial and temporal resolution, as well as accurate analysis of diffusion tracks, particularly to identify rare anomalous events such as directed motions. This study employed a Hidden Markov Model (HMM) to investigate peroxisome migration in living cells. Peroxisomes predominantly migrate in a random manner, although they do occasionally bind to the cell’s microtubular network for directed migration, which presents a significant challenge for accurate quantification. In collaboration with other researchers, the team collected diffusion tracks of thousands of peroxisomes in HEK 293 cells using high-speed two-dimensional spinning disc fluorescence microscopy.
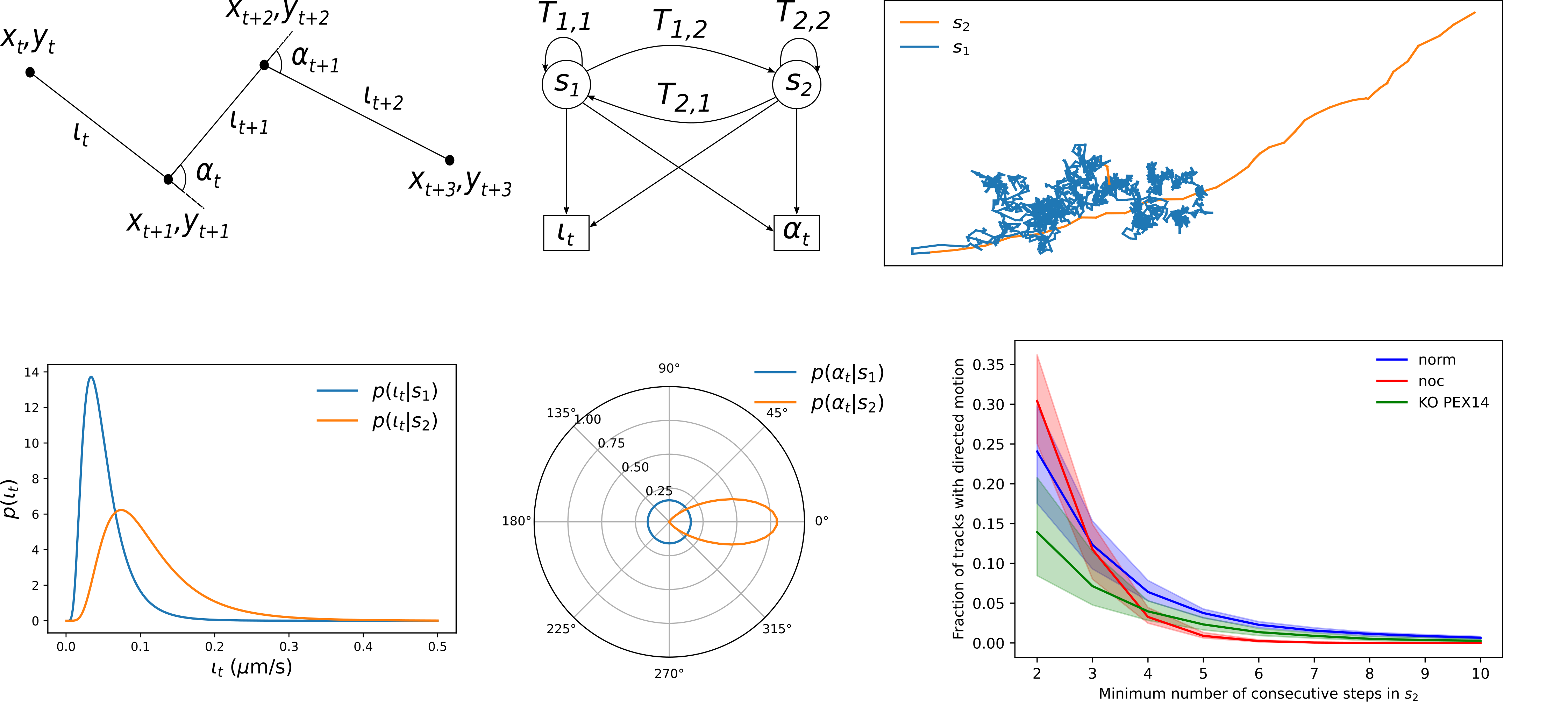
upper left: A piece of a generic track that visualizes how a series of coordinates convert to the observables instantaneous speed and turning angle (α).
bottom left: Probability density functions (pdfs) for the dimensionless instantaneous speed lt distributions of peroxisomes in the respective states s1 and s2.
upper centre: Graphical representation of the HMM with states s1 and s2, transition probabilities T1,2 and T2,1 and conversion into the parameters lt and αt bottom centre: The pdfs p(αIsi) for the turning angle and αt distributions in the respective states s1 and s2.
upper right: The two states s1 (random migration mode) and s2 (directed migration) of migration in the HMM.
bottom right: Probability density function of the minimum number of consecutive steps required for peroxisome motion under three different cellular conditions.
Comparing different cellular conditions, we show that the knockout of the peroxisomal membrane protein PEX14 leads to a decrease in the directed movement due to a lowered binding probability to the microtubule. However, it does not eradicate binding, highlighting further microtubule-binding mechanisms of peroxisomes than via PEX14. In contrast, structural changes of the microtubular network explain perceived eradication of directed movement by disassembly of microtubules by Nocodazole-treatment.
Example videos for the normal condition (https://youtu.be/LzMPtUaBxgA) and the Nocodazole treated cells (https://youtu.be/QVsfCdmSiVE). The HMM was designed with two hidden states to (1) automatically identify directed migration segments of the tracks and (2) quantify the migration properties for comparison between states and between different experimental conditions. The HMM implementation can be accessed at https://github.com/applied-systems-biology/Peroxisome_HMM.
Experimental Collaborators
- Leibniz-Institute of Photonic Technologies, Jena, Germany
- Institute of Biochemistry and Pathobiochemistry, Bochum, Germany
- MRC Translational Immune Discovery Unit, Oxford, UK





