Experiments – Whole blood infection assays
In the wet lab whole blood experiments are performed where blood samples from healthy donors or immunocompromised individuals are infected with various pathogens such as the human-pathogenic fungi Candida albicans and Candida glabrata or the bacterial pathogen Staphylococcus aureus. With the aid of FACS analysis and survival assays temporal dynamics of the host-pathogen interplay can be measured.
The project aims to improve the early detection of pathogens in bloodstream infections by analyzing immune cell behavior, specifically neutrophils.
Experimental Collaborators
- Fungal Septomics group at the Leibniz-HKI in Jena, Germany
- Institute for Hygiene and Microbiology at the Julius-Maximilians University Würzburg in Jena, Germany
State-based model
The state-based model (SBM) is a non-spatial stochastic model, which we use to simulate the interaction of different immune cell types and pathogens in a rule-based fashion. While simulating cellular interactions cells can obtain different states. For the whole blood infection assays the corresponding SBM is depicted below and is described in greater detail here.
Agent-based model
The agent-based model (ABM) is the spatial counterpart of the SBM and simulates cells as single individuals in a three-dimensional environment. The environment is modelled as a cube that resembles a section of the blood samples. Cells move and interact according to a specific rule set. The ABM can be used to estimate spatial parameters such as migration parameters for immune cells. A typical example for such a simulation can be seen below.
Agent-based model of the whole-blood infection assay with C. albicans
Here 1µl of blood is simulated comprising the immune cell types of neutrophils (blue) and macrophages (orange) and C. albicans cells in various colors depending on their current state (alive = green, dead = red, immune evasive alive = yellow, immune evasive dead = grey).
Bottom-up approach
We developed a bottom-up approach were we combined the SBM and the ABM. In this bottom-up approach non-spatial immune reaction rates are estimated using the much faster SBM. After the conversion of contact-dependent rates, such as phagocytosis rates, they can be used in the ABM. The computationally expensive ABM can then be used to estimate spatial parameters exclusively. In the future we will extend the ABM towards a hybrid ABM that – apart from the cellular layer – includes an additional molecule layer, where molecule diffusion is simulated using partial differential equations (PDE).
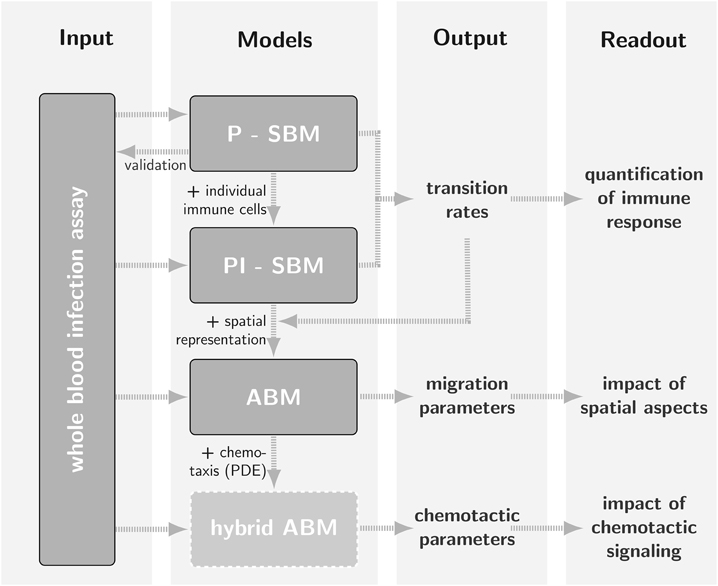
Bottom-up approach
We combined the SBM and ABM in a bottom-up fashion including self-consistent parameter conversion for efficient simulation and parameter estimation.
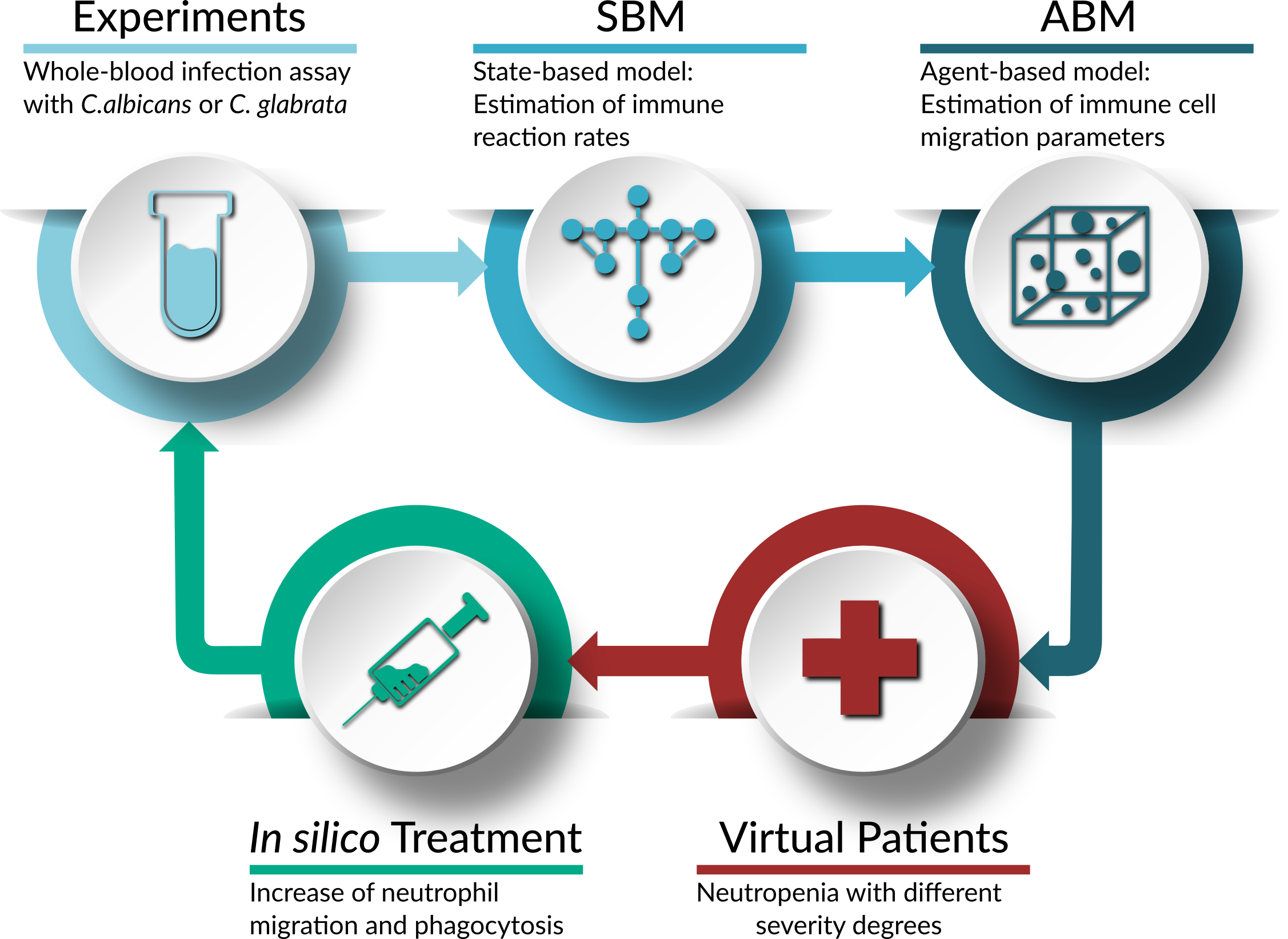
Treatment of virtual neutropenic patients
Application of the bottom-up approach to simulate fungal infections with C. albicans and C. glabrata in virtual neutropenic patients and to perform an in silico treatment of these patients.





